Our recent work in high-resolution SSNMR is an investigation of the structures of synthetic mouse prion strains. In prion diseases or transmissible spongiform encephalopathies, an aberrantly folded conformer of the prion protein (PrPSc) propagates by catalyzing post-translational misfolding of cellular prion protein (PrPC). The aberrantly folded conformers often assemble into amyloid fibrils, which coalesce into plaques, characteristic of amyloid disease. Our collaborators in the Prusiner lab have recently succeeded in recapitulating this phenomenon in vitro by converting purified, bacterially-expressed, recombinant mouse PrP into an infectious state, which has now enabled us to begin deciphering the structural basis of prion protein-based infectivity at an atomic level (1,2).
One of the more remarkable aspects of prion biology is the strain phenomenon, whereby a single protein (PrPC) can misfold into multiple infectious states with distinguishable disease phenotypes (PrPSc), and these phenotypic differences are proposed to be encoded in the physical structure of PrPSc conformers (2).
We use multi-dimensional magic-angle spinning experiments to examine structural differences among different prion strains, in the attempt to “decipher the molecular language of prion strains”. Elucidating the relationship between structure and neurological dysfunction will not only provide insight into the etiology of prion diseases such as Creutzfeldt-Jakob disease, but may illuminate the mechanism of neurodegeneration in more prevalent proteinopathies such as Alzheimer’s disease and Parkinson’s disease (3).
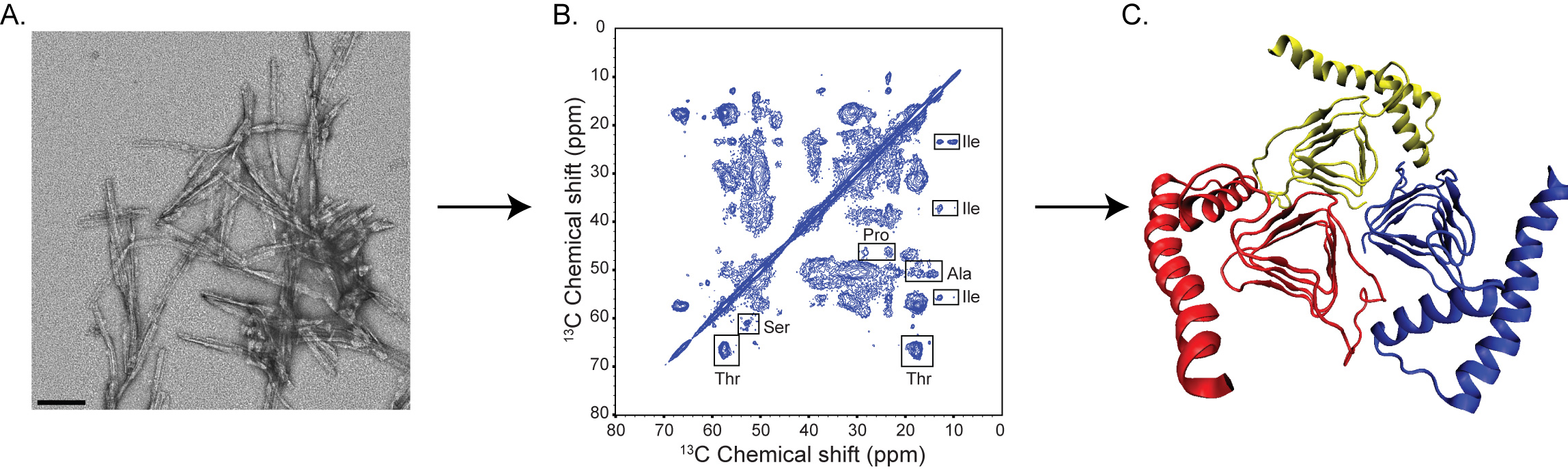
The figure above shows some of the approaches implemented and combined in elucidating the structural basis of prion disease. From left to right: (a) Negative staining electron micrograph of synthetic amyloids. The scale bar shown on the image corresponds to 100 nm. (b) 13C-13C chemical shift correlation spectrum of a uniformly-13C, 15N labeled mouse prion protein 89-230. The spectrum was acquired at 700 MHz (1H frequency). (c) Trimeric model of prions (4).
Current Members
Collaborators
David Wemmer (UC-Berkeley, Chemistry)
Stanley Prusiner (UCSF, IND)
References
[1] G. Legname, I.V. Baskakov, H.O. Nguyen, D. Riesner, F.E. Cohen, S.J. DeArmond, S.B. Prusiner. "Synthetic mammalian prions", Science, 305, 673-676 (2004).
[2] D.W. Colby, K. Giles, G. Legname, H. Wille, I.V. Baskakov, S.J DeArmond, S.B. Prusiner. "Design and Construction of diverse mammalian prion strains." Proc. Natl. Acad. Sci. USA 106, 20417-20422 (2009).
[3] S.B. Prusiner. "Cell Biology. A unifying role for prions in Neurodegenrative Diseases." Science, 336, 1511-1513 (2012).
[4] C. Govaerts, H.Wille, S.B. Prusiner, F.E. Cohen. "Evidence for assembly of prions with left-handed beta-helices into trimers." Proc. Natl. Acad. Sci. USA, 101, 8342-8347 (2004).
